The online HelitronScanner works best on a Desktop or Laptop. We recommend accessing this tool on a different device or increasing the window size to enjoy all its features.
Click the found Helitron's button to highlight its ends.
Getting Started
About the Helitron
- One remarkable feature of Helitrons is their ability to capture gene sequences, a feature that makes them of considerable potential evolutionary importance.
- It is not clear at this point how Helitrons acquire host sequences, but it is important to learn if Helitrons have the ability to trap fully functional genes and mobilize them around the genome.
- It is important to realize Helitron movement may cause changes in the flanking region, such as deletions.
- Helitrons encode proteins containing 3 conserved functional motifs that are known to be involved in bacterial and phage rolling-circle replication.
Quick-Scan
This process was created to remove obstacles for people that aren’t command-line savvy. To execute this process, copy & paste any genome sequence into the text-box and click ‘Execute’. Users are able to choose a ‘*.fa’ genome sequence file from their local machine if needed. Both methods of inputting a sequence have a limit of 1 Mb file size. If a user has a file that is larger than the limit, we recommend the user to download a copy of the Helitron-Scanner.
Once the process has completed a sequence containing the Helitron will be outputted.The 5' and 3' ends of the sequence will also be highlighted with the users chosen color. If the scan produced no results a display message will be posted. Users also have the ability to download results to a file name and extention of their choosing. The default download file name for the Quick-Scan process is ‘output_hel.fa’.


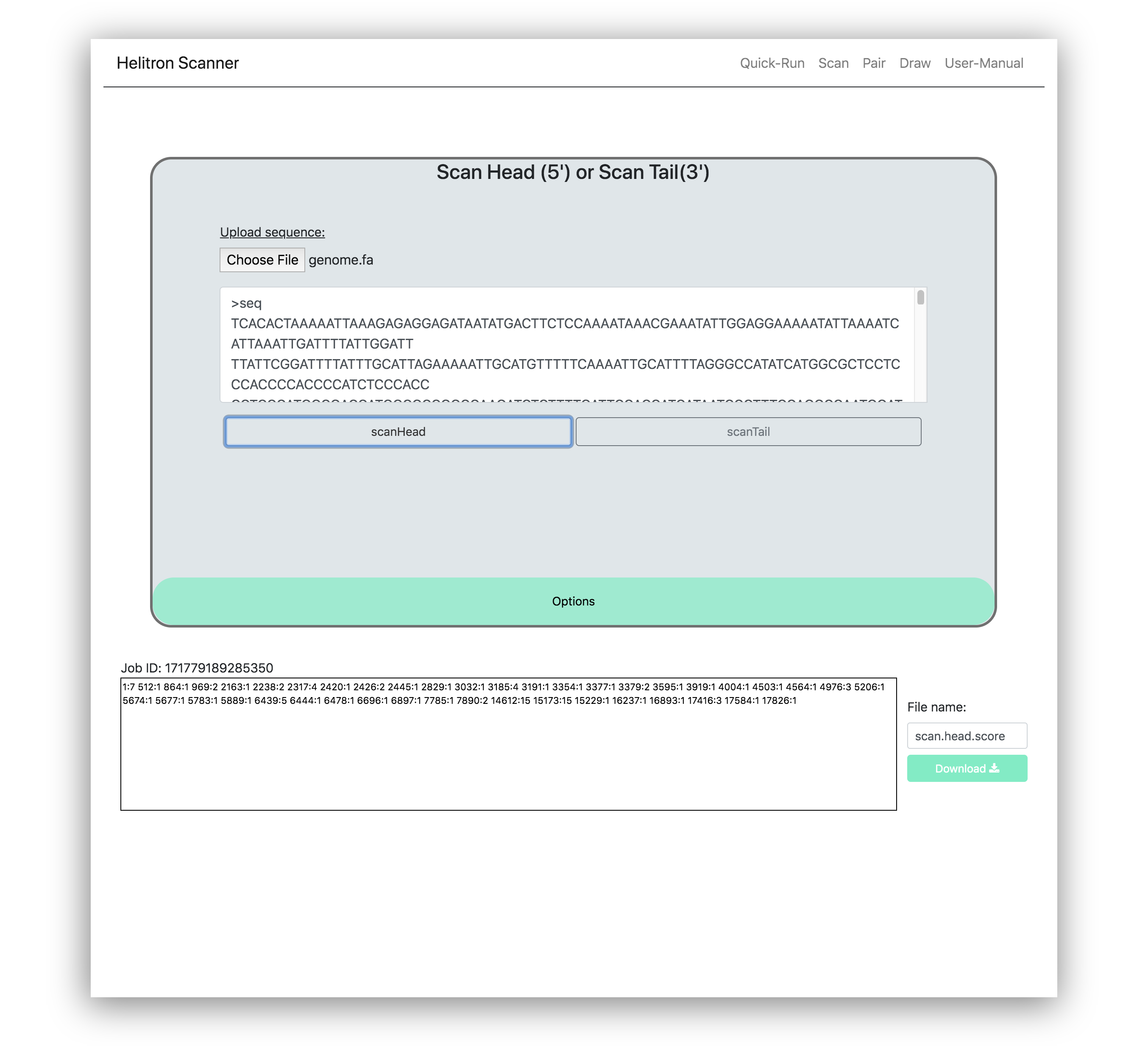
Scan
This process scans for Helitrons in the 5' (scanHead) and 3'(scanTail) termini respectively. To utilize these functions, upload or paste your sequences into the interface. Then click scanHead or scanTail for your desired termini scan. An example of a sequence that has been scanned in the 5' termini can be viewed below.
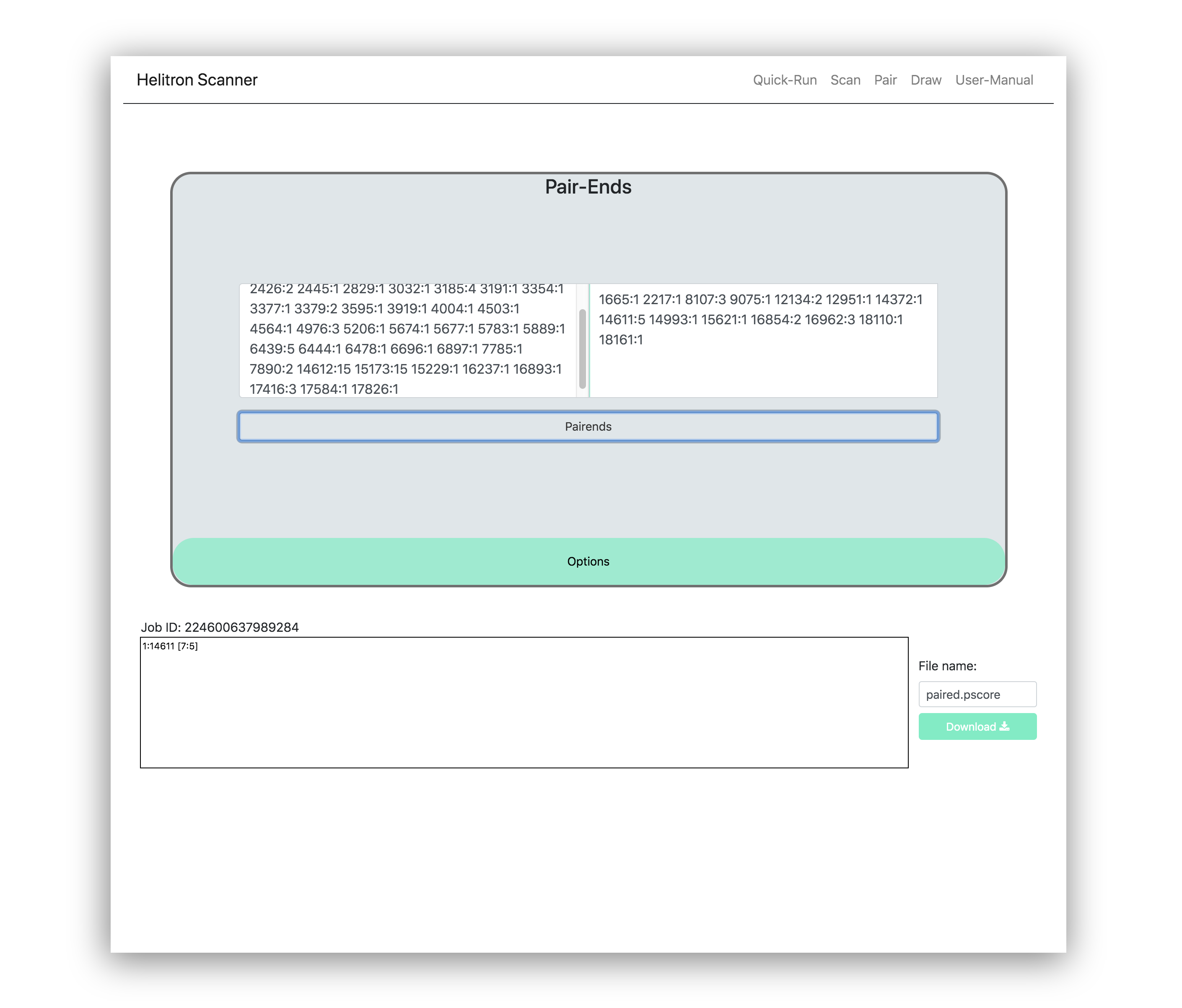
Pair
This process takes the location scores or output from both Helitron ends and pairs them to create coordinates for the Helitron in the original sequence. To utilize this function, the user must copy & paste the output from the 5' and 3' termini from the scan tab found in the top right of this web-application.
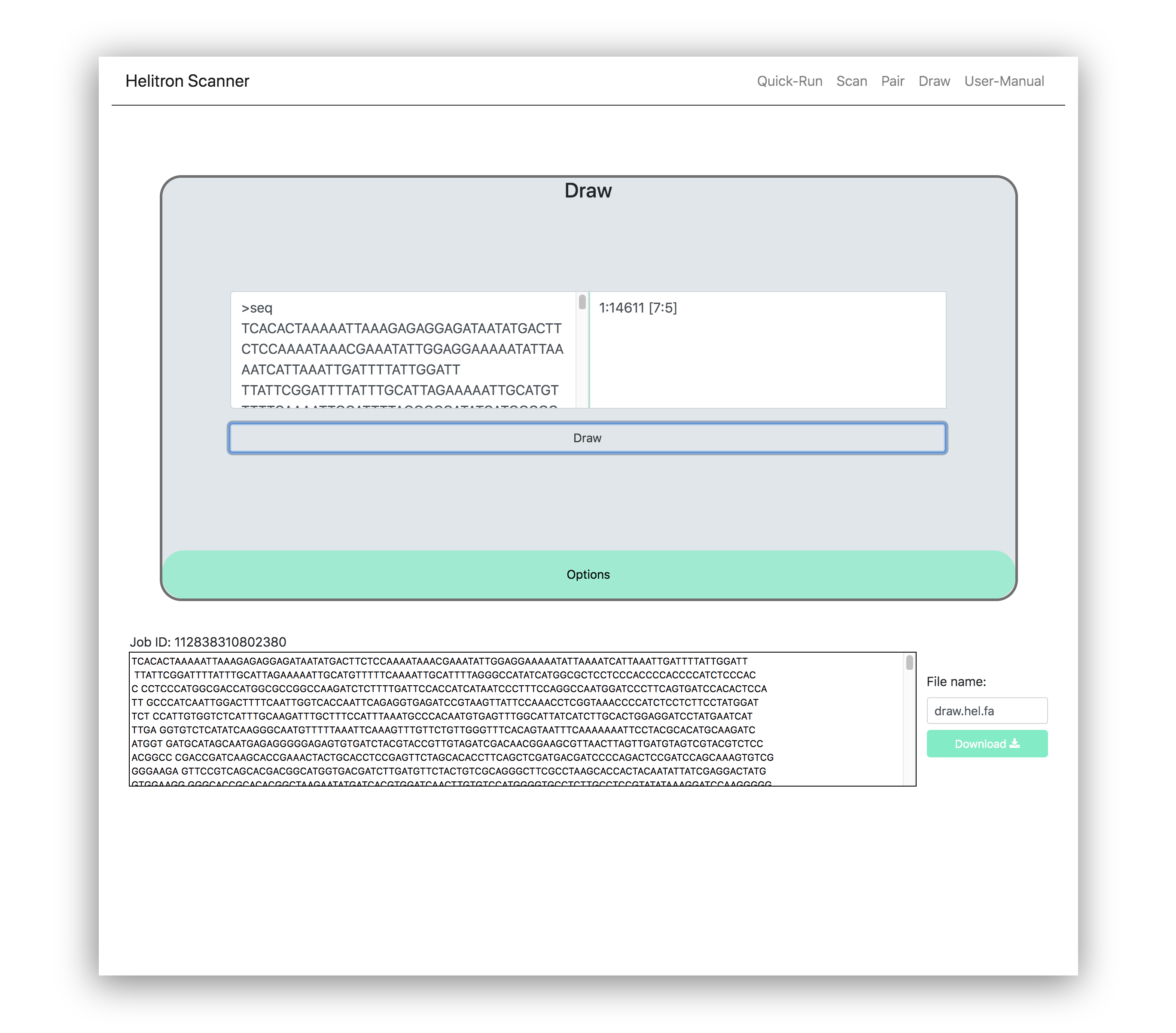
Locate
This process takes the pair-ends score or output generated from the pair-ends function, found in the Pair tab, and locates the Helitron in the related regions from the original input file.
Browser compatibility:
OS
Version
Chrome
Firefox
Microsoft Edge
Safari
MacOS
Catalina
78.0.3904.108
71.0
44.18362.449.0
13.0
Windows
10
78.0.3904.108
71.0
44.18362.449.0
n/a
Linux
Debian
78.0.3904.108
71.0
n/a
n/a
Citation
- Wenwei Xiong & Chunguang Du (2014) Mining hidden polymorphic sequence motifs from divergent plant helitrons, Mobile Genetic Elements, 4:5, 1-5, DOI: 10.4161/21592543.2014.971635
- Chunguang Du & Jason Caronna & Limei He & Hugo K Dooner (2008) Computational prediction and molecular confirmation of Helitron transposons in the maize genome, BMC Genomics, 9:51, DOI: 10.1186/1471-2164-9-51
- Xiong, W., L. He, J. Lai, H.K. Dooner, C. Du. 2014. HelitronScanner uncovers a large overlooked cache of Helitron transposons in many genomes. Proc Natl Acad Sci USA Vol. 111: 10263-10268
Resources used:
This website uses bootstrap for front-end styling.Bootstrap Tour was used to help new users with a touring feature. JQuery and Font Awesome were also utilized for javascript simplicity and icons, respectively.